PDFLINK |
Exciting Coding Problems for DNA-based Storage Systems
Communicated by Notices Associate Editor Richard Levine
Introduction to DNA Storage
The potential for using macromolecules for ultra-dense storage was recognized as early as in the 1960s, when the celebrated physicist Richard Feynman outlined his vision for nanotechnology in the talk “There’s plenty of room at the bottom” Fey92. DNA lends itself to implementations of non-volatile recoding media of outstanding integrity due to its unique properties; among them are its extremely high capacity and stability. Furthermore, the technologies for synthesizing (writing) artificial DNA and for sequencing (reading) DNA have reached unprecedented levels of efficiency and accuracy Bd14Eis14. Thus, DNA-based storage systems are predicted to materialize in the near future.
Storing Digital Data in DNA
DNA consists of four types of nucleotides: adenine (A), cytosine (C), guanine (G), and thymidine (T). A single DNA strand, also called an oligonucleotide (oligo), is an ordered sequence of the nucleotides. The beginning and end of a DNA strand are distinguishable as they are chemically different. Each strand starts with the so-called 3’ nucleotide end and it ends with the 5’ nucleotide end. DNA strands can be synthesized chemically and modern DNA synthesizers can concatenate the four DNA nucleotides to form almost any sequence. This process enables the strands to store digital data. The data can be read back with common DNA sequencers, where the most popular ones use DNA polymerase enzymes or nanopore-based sequencing and are referred to as sequencing by synthesis Bd14.
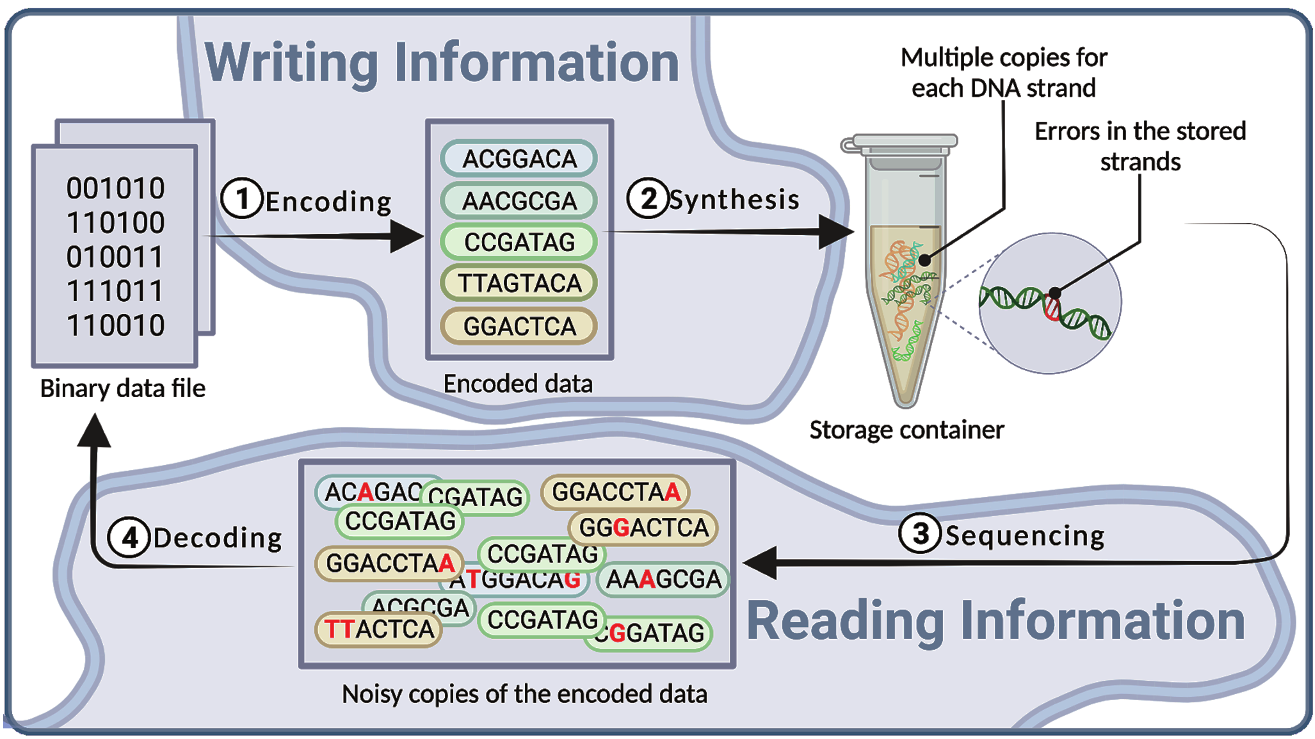
A DNA storage system typically consists of three important entities; see Fig. 1. The first is a DNA synthesizer which produces the strands that encode the data to be stored in DNA. In order to produce strands with an acceptable error rate, the length of the strands is typically limited to no more than 250 nucleotides CCS19. Additionally, every strand has millions of synthesized copies. The second part is a storage container with compartments that store the DNA strands, however unordered. Lastly, a DNA sequencer reads the strands and transfers them back to digital data. The encoding and decoding stages are two external processes to the storage systems which encode the binary user data into strands of DNA in such a way that even in the presence of errors (the nucleotides in red in Fig. 1), it will be possible to revert back to the original binary data of the user.
DNA as a storage system has several attributes which distinguish it from any other storage system. First, the synthesis and sequencing processes introduce a unique error behavior that is mostly dominated by deletions, substitutions, and insertions of symbols. Second, the strands are not ordered and thus it is not possible to know the order in which they were stored. Usually, this problem can be overcome by using indices, that are stored as part of the strand, and this limitation already imposes the capacity of DNA storage to be strictly less than 2 bits per nucleotide. Third, every strand has millions of copies and this inherent redundancy should be taken advantage of during decoding.
Previous Work
Storing data in DNA is not a new idea as it was already mentioned by Richard Feynman more than 60 years ago. Yet, it took more than 50 years to conduct the first large scale experiments that demonstrated the potential of in vitro DNA storage. Church et al. stored 643 KB of data CGK12 and Goldman et al. accomplished the same task for a 739 KB message GBC13. However neither of these groups recovered the entire message successfully since they did not use appropriate coding solutions to correct the typical errors in DNA. Since then, additional academic groups and companies have been developing feverishly new ideas to efficiently store digital data in DNA while reducing the cost; for an example see BOS21 and MTK21CCS19 for an extensive summary of this progress and future forecast.
Error Correction in Storage Systems
Digital data is stored in computers as vectors (words) over some alphabet . The process of storing digital data starts with setting an injective function on the desired data that maps it into coded vectors (codewords) over . This mapping is necessary since no storage system is error-free. Furthermore, the design of the mapping depends mostly on the type of errors in the storage system and their distribution, which varies between different storage media. Therefore, in order to design a storage system which is resilient to errors, one should consider the characteristics of the considered medium to create a suitable mapping, which is referred to as an error-correcting code. More precisely, in order to overcome up to some errors within a stored codeword , we first need to consider the set of all erroneous versions of the codeword that can be obtained from it by introducing up to errors to . This set of erroneous words is referred to as the radius- error ball of , and is denoted by . Thus, an error-correcting code that can correct up to errors, is a set of codewords of a certain length, whose radius- error balls are mutually disjoint. In other words, the stored codewords can be uniquely identified by each of their erroneous versions.
There are numerous families of error-correcting codes, which vary mostly by the type of errors they correct, the error-correction guarantees, and the encoding/decoding complexity. For some of the codes the goal is to correct the desired errors in the worst case, i.e., any errors (or less) are guaranteed to be corrected and can be a constant or a function of the word’s length. On the other hand, other codes aim to correct these errors with high probability. Additionally, some codes are designed to address the probabilistic case, in which each symbol is erroneous with some fixed error probability.
In almost all of the conventional storage media, such as tapes, hard-disk drives, and solid-state drives, the errors that occur are symbol erasures and/or symbol substitutions, for which it is known that the size of the error balls depends solely on the number of errors and not on the codeword itself, i.e., the size of is the same for any word of the same length. Due to the prevalence of these errors, there are many families of error-correcting codes and other coding techniques that are targeted to correct substitutions and erasures such as RS, LDPC, BCH, polar, and convolutional codes LC04.
Mathematical and Computational Problems of DNA Storage
As mentioned above, the microscopic world in which the DNA molecules reside induces error patterns that are fundamentally different from their digital counterparts. These properties lead to the following exciting problems.
1. Error-correcting codes for DNA storage systems
The problem of constructing codes that correct insertions, deletions, and substitutions, which are referred to as edit errors, is one of the more intriguing challenges in the area of coding theory. While symbol substitutions/erasures effect only the defected symbol(s), insertion and deletion of symbols can generate undesired indentation of the correct symbols in the word, and can also affect its length. Moreover, the size of error balls that consider deletion errors depends not only on the radius , but also on the inspected word. These sizes are known only when is very small. The first family of such codes was proposed by Varshamov and Tenengoltz in 1965 VT65 and are referred to as VT codes. In fact, Varshamov and Tenengoltz proposed these codes to correct a single asymmetric error, in which only a 0 can change to be a 1. A year after Levenshtein showed that this family of codes can correct a single insertion or a single deletion Lev65. Levenshtein also showed that a code can correct deletions if and only if it can correct any combination of insertions and deletions Lev65. A length- VT code is defined by an integer and consists of all binary vectors that satisfy the following constraint,
It is known that for any , the code is a perfect single-deletion-correcting code, that is, the union of the disjoint radius- error (deletion) balls of the words in covers the entire space of vectors of length . It is interesting to mention that even though the VT code is perfect for any , since the size of the radius- error balls varies, the number of words in also varies for different values of . Furthermore, the case yields the largest, smallest VT code, respectively; namely for any , . It is still an open question whether there exists a length- single-deletion-correcting code , such that , while it is commonly conjectured that the code is strictly optimal for all values of Slo02.
The error behavior of the DNA storage channel renewed the interest in studying the deletion and insertion channels and synchronization channels in general. Studying and analyzing these channels is a notoriously hard problem in the coding and information theory communities, which are far from being fully solved. Nevertheless, these channels introduce intriguing coding and information-theoretic questions and were studied extensively in the literature. We refer the reader to an excellent survey for more information on these problems and other information-theoretic problems which are related to DNA data storage SH22.
2. The DNA clustering problem
In current synthesis technologies, every synthesized DNA strand has millions of copies, which leads to inherent redundancy that is unique to DNA storage systems. This property can be utilized to enhance the error-correction capabilities. However, due to the unordered nature of the medium, in order to take advantage of this inherent redundancy, one should first order and cluster all of the received erroneous copies of the synthesized strands. This additional step is not common among other digital storage media and thus it introduces a new set of interesting research problems. The sequencing process results with an unordered set of millions to billions DNA sequences, which are all the noisy copies of the designed DNA strands that encode the data. In the DNA clustering problem, the goal is to partition the set of read strands into small sets, which are referred to as clusters, such that in each cluster there are only copies that originated from the same designed DNA strand.
Previous works attempted to tackle this problem by either designing clustering algorithms that consider the edit distance between the strands or by encoding the information in a way that facilitate the clustering procedure. However, the DNA clustering problem is still far from being solved. Most of the existing solutions do not perform well under high error rates, which are prevalent among new emerging cheaper/faster synthesis and sequencing technologies. Additionally, the performances and hardware requirements of the currently existing algorithmic-based methods cannot scale to meet the potential storage volume of future DNA-based systems. Even though combining coding techniques can theoretically improve the latency and save physical and logical resources, such solutions are either relatively naive and non-optimal (e.g., requires large redundancy) or are based on unrealistic assumptions (e.g., only specific error patterns) which make them inapplicable in real systems.
3. The sequence reconstruction problem
After the clustering step, each cluster consists of the read strands, which are noisy copies of one of the synthesized strands and should be used for its recovery. In this problem the goal is to estimate the designed DNA strand (with zero or small error probability), from a given cluster of its noisy copies. One of the challenges in the DNA storage channel is that we do not necessarily have control on the size of the clusters, and it is very likely that this size is significantly smaller than the minimum size that guarantees a unique decoding of the designed DNA strands. Mathematically speaking, a DNA reconstruction algorithm is a mapping which receives some noisy copies as an input and produces , an estimation of , and the goal is to minimize the distance between and . The problem of estimating a sequence from a set of its erroneous versions is also related to the trace reconstruction problem BKKM04 and the sequence reconstruction problem Lev01. In these problems the goal is to determine the required number of distinct copies to ensure a unique decoding with high probability or in the worst case.
4. Constrained codes
To reduce the error rates of the synthesis and the sequencing processes, there are several design constraints that are unique to DNA storage and should be enforced on the designed DNA strands. A constrained code is defined as a set of codewords that satisfy some given design constraints. Some of these constraints are relevant to the majority of the technologies that are being used nowadays and two of the main ones are: 1. The run-length constraint, in which the goal is to limit the number of identical consecutive symbols in any of the designed DNA strands (this number is typically between 3 and 5). 2. The GC-content balanced constraint, in which the quantity of the ‘G’ and ‘C’ symbols in each DNA strand should be roughly balanced, for example between 45% and 55%. The first constraint is an example of a local constraint, and the second is considered a global one. While dealing with either one of these constraints is a doable task, combining both the local and the global constraints is a more challenging task with significantly less knowledge. Additionally, new emerging synthesis and sequencing technologies may introduce new desired constraints. Even though constrained coding has been well studied, it is still unknown how to design efficient codes for all constraints and combine them with error-correcting codes.
The behavior and properties of these four problems depend on the synthesis and the sequencing technologies that are being used SOS21. Recently, a few emerging synthesis technologies were suggested, while each such a technology presents new error behavior that should be considered and addressed separately LKG19AVA19. Any such new technology opens the door to a variety of intriguing and exciting theoretical problems and challenges.
Conclusion
When designing new coding schemes for DNA-based storage systems, all of the properties of the DNA storage system that were mentioned in this review should be considered. That being said, the current implementations of DNA-based storage systems are very limited and are not fully optimized to address the unique pattern of errors which characterize the synthesis and the sequencing processes. Hence, to maintain reliability in reading and writing, new coding schemes must be developed. Moreover, since DNA storage aims to address orders of magnitude more data compared to current technologies, it is extremely important for new coding schemes and algorithms to be scalable and efficient. DNA storage generates new exciting and challenging theoretical problems to our community. The success of such solutions is crucial due to the never-ending growth of data and the foreseeable inability to store it.
A DNA-based storage system. 1) Encoding of the binary information. 2) Synthetic DNA strands that encode the data are synthesized using a DNA synthesis technology. 3) Sequencing a sample from the DNA storage container using a DNA sequencer; The number of copies for each strand can vary and some strands may have zero or very small number of copies. 4) Lossless decoding to retrieve the binary data.
References
- [AVA19]
- L. Anavy, I. Vaknin, O. Atar, R. Amit, and Z. Yakhini, Data storage in DNA with fewer synthesis cycles using composite DNA letters, Nature Biotechnology 37 (2019), no. 10, 1229–1236.
- [BOS21]
- D. Bar-Lev, I. Orr, O. Sabary, T. Etzion, and E. Yaakobi, Deep DNA storage: Scalable and robust DNA storage via coding theory and deep learning, Preprint, arXiv:2109.00031, 2021.
- [BKKM04]
- T. Batu, S. Kannan, S. Khanna, and A. McGregor, Reconstructing strings from random traces, Proceedings of the Fifteenth Annual ACM-SIAM Symposium on Discrete Algorithms, ACM, New York, 2004, pp. 910–918. MR2290981Show rawAMSref
\bib{BKK04}{article}{ author={Batu, Tu\v {g}kan}, author={Kannan, Sampath}, author={Khanna, Sanjeev}, author={McGregor, Andrew}, title={Reconstructing strings from random traces}, conference={ title={Proceedings of the Fifteenth Annual ACM-SIAM Symposium on Discrete Algorithms}, }, book={ publisher={ACM, New York}, }, date={2004}, pages={910--918}, review={\MR {2290981}}, }Close amsref.✖ - [Bd14]
- H. P. J. Buermans and J. T. den Dunnen, Next generation sequencing technology: Advances and applications, Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease 1842 (2014), no. 10, 1932–1941.
- [CCS19]
- D. Carmean, L. Ceze, G. Seelig, K. Stewart, K. Strauss, and M. Willsey, DNA data storage and hybrid molecular–electronic computing, Proceedings of the IEEE 107 (2019), no. 1, 63–72.
- [CGK12]
- G. M. Church, Y. Gao, and S. Kosuri, Next-generation digital information storage in DNA, Science 337 (2012), no. 6102, 1628–1628, available at https://www.science.org/doi/pdf/10.1126/science.1226355.
- [Eis14]
- M. Eisenstein, Next generation sequencing technology: Advances and applications, Nature Biotechnology 38 (2014), no. 10, 1113–1115.
- [Fey92]
- R. P. Feynman, There’s plenty of room at the bottom, Journal of Microelectromechanical Systems 1 (1992), no. 1, 60–66, DOI 10.1109/84.128057.
- [GBC13]
- N. Goldman, P. Bertone, S. Chen, C. Dessimoz, E. M. LeProust, B. Sipos, and E. Birney, Towards practical, high-capacity, low-maintenance information storage in synthesized DNA, Nature 494 (2013), no. 7435, 77–80.
- [LKG19]
- H. H. Lee, R. Kalhor, N. Goela, J. Bolot, and G. M. Church, Terminator-free template-independent enzymatic DNA synthesis for digital information storage, Nature Communications 10 (2019), no. 1, 1–12.
- [Lev01]
- V. I. Levenshtein, Efficient reconstruction of sequences, IEEE Trans. Inform. Theory 47 (2001), no. 1, 2–22, DOI 10.1109/18.904499. MR1819952Show rawAMSref
\bib{LEV-Rec}{article}{ author={Levenshtein, Vladimir I.}, title={Efficient reconstruction of sequences}, journal={IEEE Trans. Inform. Theory}, volume={47}, date={2001}, number={1}, pages={2--22}, issn={0018-9448}, review={\MR {1819952}}, doi={10.1109/18.904499}, }Close amsref.✖ - [Lev65]
- V. I. Levenshtein, Binary codes capable of correcting deletions, insertions, and reversals, Soviet Physics Dokl. 10 (1965), 707–710. MR0189928Show rawAMSref
\bib{LEV66}{article}{ author={Levenshtein, V. I.}, title={Binary codes capable of correcting deletions, insertions, and reversals}, journal={Soviet Physics Dokl.}, volume={10}, date={1965}, pages={707--710}, issn={0038-5689}, review={\MR {0189928}}, }Close amsref.✖ - [LC04]
- S. Lin and D. J. Costello, Error control coding, second edition, Prentice-Hall, Inc., USA, 2004.
- [MTK21]
- K. Matange, J. M. Tuck, and A. J. Keung, DNA stability: a central design consideration for DNA data storage systems, Nature Communications 12 (2021), no. 1, 1–9.
- [SOS21]
- O. Sabary, Y. Orlev, R. Shafir, L. Anavy, E. Yaakobi, and Z. Yakhini, SOLQC: Synthetic oligo library quality control tool, Bioinformatics 37 (2021), no. 5, 720–722.
- [SH22]
- I. Shomorony and R. Heckel, Information-theoretic foundations of DNA data storage, 2022.
- [Slo02]
- N. J. A. Sloane, On single-deletion-correcting codes, Codes and designs (Columbus, OH, 2000), Ohio State Univ. Math. Res. Inst. Publ., vol. 10, de Gruyter, Berlin, 2002, pp. 273–291, DOI 10.1515/9783110198119.273. MR1948149Show rawAMSref
\bib{Sloane}{article}{ author={Sloane, N. J. A.}, title={On single-deletion-correcting codes}, conference={ title={Codes and designs}, address={Columbus, OH}, date={2000}, }, book={ series={Ohio State Univ. Math. Res. Inst. Publ.}, volume={10}, publisher={de Gruyter, Berlin}, }, date={2002}, pages={273--291}, review={\MR {1948149}}, doi={10.1515/9783110198119.273}, }Close amsref.✖ - [VT65]
- R. R. Varshamov and G. M. Tenengoltz, A code for correcting a single asymmetric error, Automatica i Telemekhanika 26 (1965), no. 2, 288–292.
Credits
Figure 1 is courtesy of the authors and was created with BioRender.com.
Photo of Daniella Bar-Lev is courtesy of Daniella Bar-Lev.
Photo of Omer Sabary is courtesy of Omer Sabary.
Photo of Eitan Yaakobi is courtesy of Eitan Yaakobi.




