Mathematics and the Genome: Near and Far (Trees)
6. Near and Far (Trees)
If one wants to get a sense of family history, one draws a family tree. Scientists who want to study the relatedness, history or evolution of objects also draw trees and use a body of mathematical knowledge to do so. This body of knowledge includes a lovely theory of relating trees to distance. Suppose that for every pair of the objects it is possible to define a distance between these pair of objects. For example, this distance might measure systematic changes that occurred to transform a gene in one species to a related gene in another species. More specifically, imagine that one has 4 DNA sequences a, b, c, and d whose edit distances are as shown in the matrix (table) below:

Can one label the leaves of a tree (the vertices where there is one edge) structure such that the sum of the weights along the path joining these leaves is given by the entry in the table? For example, the unique path (trees always have a unique path between any pair of their vertices, which is one of the reasons they have nice mathematical properties) between a and c has weights which sum to 9 (3 + 2 + 4) which is the entry in row a column c in the matrix above.

This tree can be said to represent the matrix because all the distances in the matrix correspond to the sums of the weights on the paths. There is a nifty theorem that characterizes when a distance matrix can be represented by a tree which has only 1-valent (i.e. the leaves of the tree) and 3-valent vertices, as does the tree above, and where the rows of the distance matrix are used to label the 1-valent vertices of the tree. This theorem states that if one looks at any four objects x, y, z, w a representation is possible if and only if the distance xy plus distance zw, the distance xz plus distance yw, and the distance xw plus distance zy are not distinct and the two larger numbers one gets are equal. In the example above we get 13, 17, and 17 for these three numbers. The condition is known as the 4-point condition. This is a theorem of P. Buneman. There is also a similar 3-point condition that guarantees that the distance matrix is representable by a special type of tree called an ultrametric tree. The diagram below shows an example.
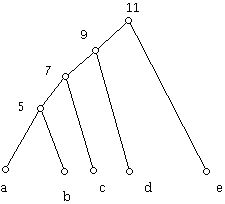
The distance between the manuscripts represented by the leaves is the number on the lowest internal vertex of the tree that lies above these leaves. For example, the distance between b and d is 9. This number can be thought of as the number of years back in the past where b and d had a common ancestor. The distance between all the manuscripts is shown in the matrix (table) below:

The three-point condition says that a distance matrix can be represented in this way by a tree if and only if for any three objects x, y, and z the distances between them is such that the two largest are equal.
Unfortunately, with real data there is little hope that it will obey very special conditions of this kind. Thus, researchers have looked at a wide variety of methods to compute a best tree representing the distances. There is also considerable work in finding a consensus tree for reconciling trees that may have been arrived at for the same objects from different points of view. For example, one might have trees developed on the basis of morphological data and other trees based on genetic data. This research has assisted biologists in getting understanding while, at the same time, yielding many interesting new mathematical methods and insights.
-
Introduction
-
Mathematics and Classical Genetics (The Early Days)
-
Mathematics and Classical Genetics (1900-1953)
-
Molecular Genetics (1953-Present)
-
Near and Far (Strings)
-
Near and Far (Trees)
-
The Wider Picture and the Future
-
References
Welcome to the
Feature Column!
These web essays are designed for those who have already discovered the joys of mathematics as well as for those who may be uncomfortable with mathematics.
Read more . . .
Feature Column at a glance